With more and more companies being serious about their data, file sizes are now larger than they used to be. While most of our laptops still have 8GB of RAM, we get out of memory errors when we try to open files larger than memory. Instead of adding more memory to your system (or using a cloud service), we can have another way to deal with this situation.
Some people might be confused about these two terms:
**Storage ** is refer to HDD/SSD
**Memory ** is refer to RAM
In this blog, I would introduce methods for interacting with large data using Linux and Python.
Environment Preparation
The dataset I used for this experiment is the CORD-19 dataset.
At this snapshot, the cord_19_embeddings_2022-02-07.csv is around 13G and the environment I used has only 12G memory. You can check your computer memory configuration using free -h. And check the file size using ls -lh. Also check the disk size using df -h.
Bonus: You could use
watchto watch the changes from time to time.
All the code used is in the following Colab notebook:
Handling with Linux Command
GNU Coreutils
Command: cat/head/tail/more/less
The Linux terminal has always been a powerful tool for working with files. It provides powerful command-line tools to manipulate files. The following commands are from GNU Coreutils, which you can find on almost every Linux system. When working with large files, GUI interface programs can have difficulty opening the file. In this case, command-line tools can come in handy.
| Command | Description |
cat myfile.csv | View the entire file |
head/tail myfile.csv | View the head/tail of the files |
more/less myfile.csv | View the file page by page |
Command: split
#!/bin/bash
# n x 14MB files
split cord_19_embeddings_2021-05-31.csv
# n x 1G files
split -b 1G cord_19_embeddings_2021-05-31.csv
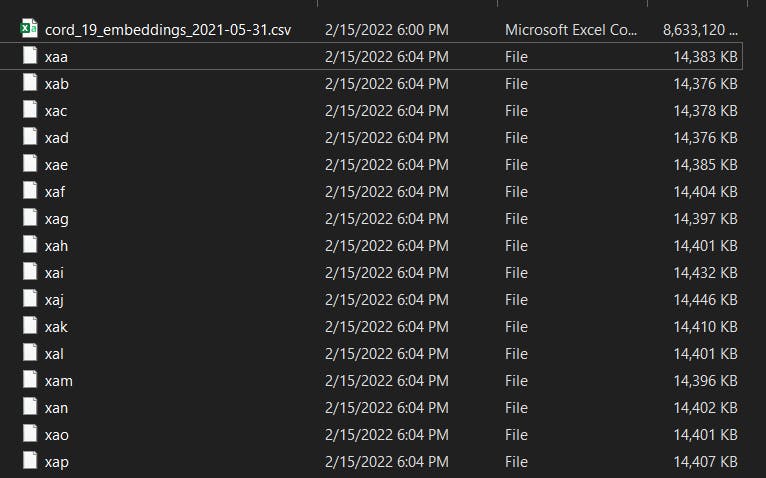
The image above shows the result when you just run the command split file.csv. The file is actually a plain text file. You can still rename the files with extra extension and you can now open the .csv file with Excel. To make life easier, we can ask the tool to rename it for us. Just add some arguments as below:
# n x 1G files with -Digit.csv as suffix
split -b 1G -d --additional-suffix=.csv cord_19_embeddings_2021-05-31.csv cord_19_embeddings
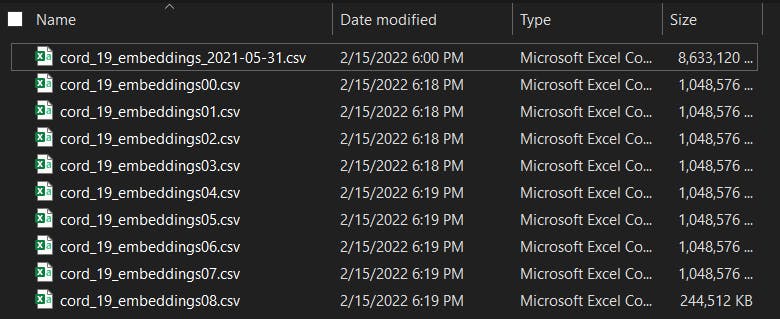
To fully understand the meaning of the argument, check the official documents: Coreutil: Split a file into pieces.
To count all the line of the csv files: wc -l *.csv
For Windows User
I strongly recommend that you install the ubuntu subsystem in Windows 10. If you want to stick with Microsoft, try exploring PowerShell or Batch scripting on Windows. After installing Linux Subsystem, in Windows run:
ubuntu run [your-linux-command]
Additional Reading:
Handling with Python
Using Pandas
Our beloved Pandas is the most popular data analysis tool. It is an open-source Python library based on the NumPy library. By nature, it is not supposed to handle such a large amount of data. Pandas is "well-known" for a certain design problem, like, Pandas is single-threaded. When dealing with datasets as large as the memory, it can be trickier to deal with.
Using chunking might be a way out. Chunking means that we do not load the entire data into memory. Instead, we want to load one part of the data, process it, then load another part, and so on. In the input argument to pandas.read_csv(), you can specify chunk size data at load time. Below is the performance comparison:
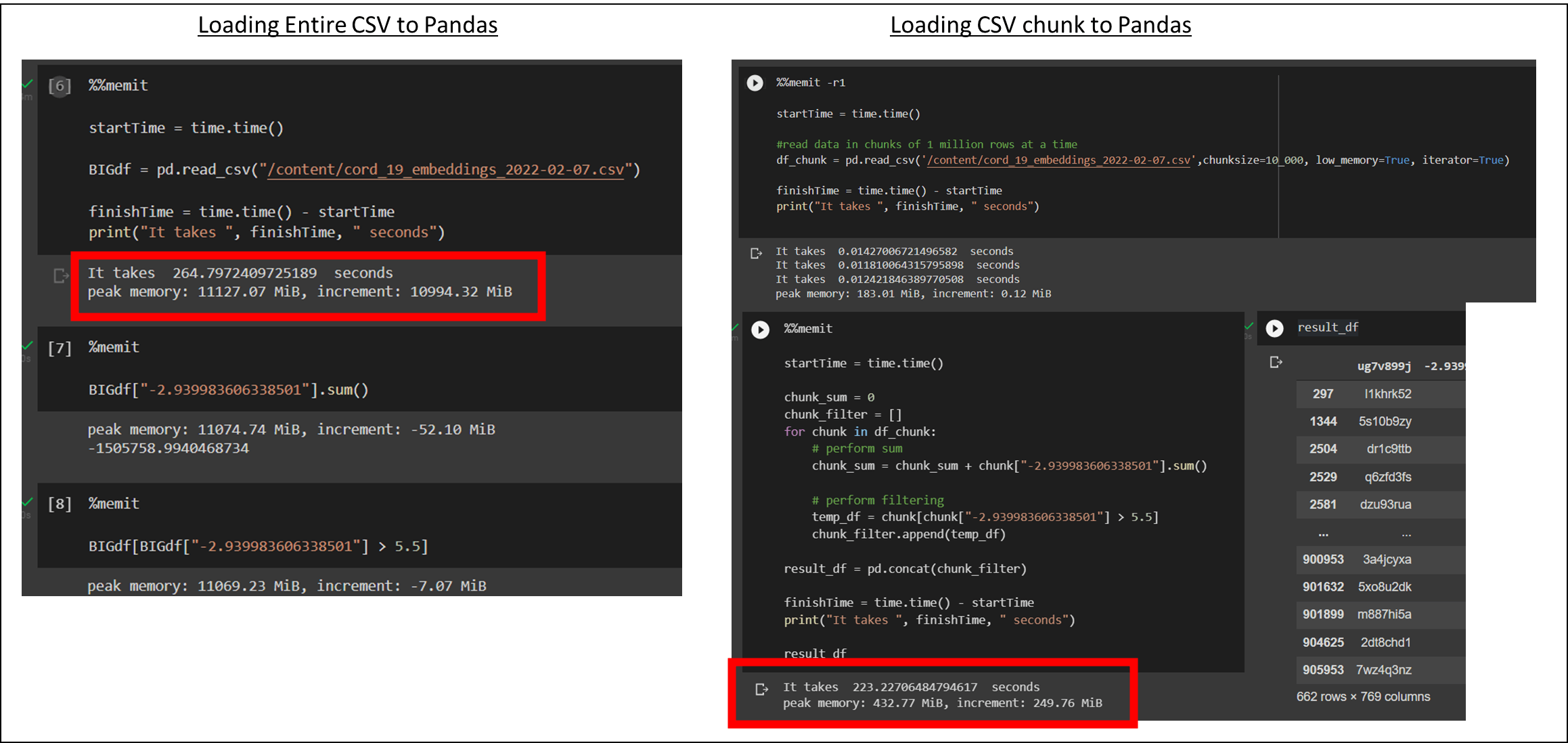
| Performance | W/o Chunk | w/ chunk |
| Time | 264s | 223s |
| Peak Memory | 11127.07MiB | 432.77MiB |
| Increment Memory | 10994.32MiB | 249.76MiB |
The time required for running the query is almost the same because they are loading the same amount of data using a single thread. But when it comes to memory, it makes a big difference. The memory usage remains almost constant, which is manageable for us, and technically the data size can scale to infinity! The concept is just like doing stream processing.
df_chunk is an iterator object which we can use other advance Python tools like itertools to further speed up the process of analytics. Here I just simply use a for loop to process the result.
Additional Reading:
Using Dask

Dask is an Open Source library for parallel computing written in Python. It is a dynamic task scheduler and a "big data" library that extend the capability of NumPy, Pandas. The design logic behind Dask would be by writing the task using high level collections with Python, and Dask will generate task graphs that can be executed by schedulers on a single machine or a cluster. As a result, Dask is a lazy evaluation language. We write our pipeline and run .compute to really execute the program. This ensures that some tasks can be executed in parallel.
The syntax of the Dask Dataframe is more or less the same as Pandas. The main difference would be importing Dask library with import dask.dataframe as dd, and running .compute to obtain the result. Friendly reminder, we should avoid calling compute repeatedly.
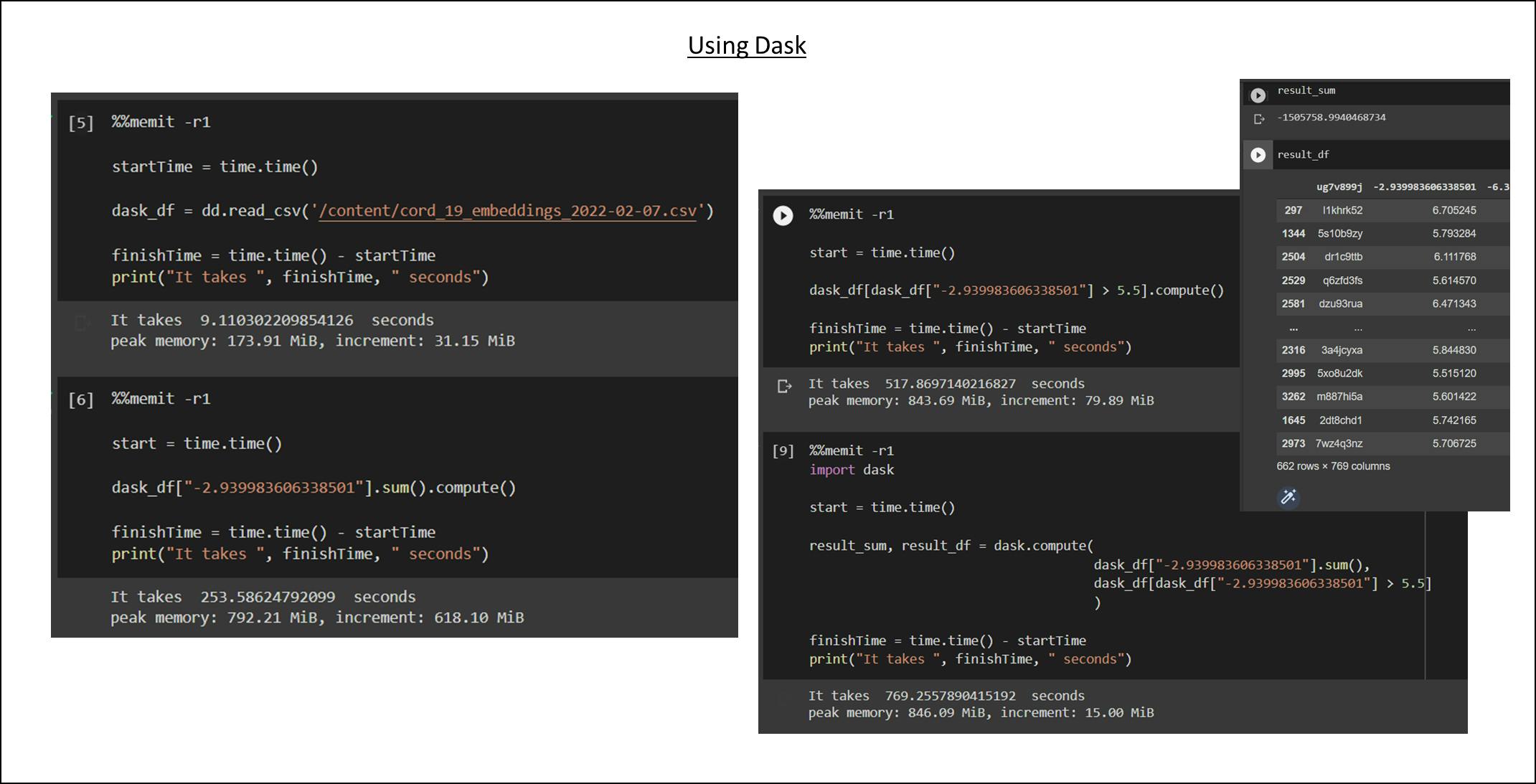
| Performance | Loading CSV | Compute Sum | Filter Dataframe | Compute Both | | ----------- | ----------- | ----------- | | Time | 9s | 183.91s | 158.58s | 769s | | Peak Memory | 173.91 MiB | 792.21 MiB | 843.69 MiB | 846.09 MiB | | Increment Memory | 31.15 MiB | 618.10 MiB | 17.1 MiB | 74.87 MiB|
Loading csv is so fast because Dask only loads data when we need to get query results. There is some technique behind calculating the sum while filtering requires reading the entire data. That's why it takes more time to filter then to compute a sum. This is just the tip of the iceberg for using Dask, feel free to check out the Dask Document.
Explore the possibility
Usually, when dealing with large files, we harness the power of distributed computing. Like if we can load the file into HDFS, we can leverage the Hadoop ecosystem like Apache Spark to process really big data. Dask provides Dask.distributed for distributed computing in Python. Coiled provides extra cloud computing power for Dask users. Ray is a low-level framework for parallelizing Python code across processors or clusters. It is a relatively new package that many big players are adopting to using it. Ray has a managed cloud offering called Anyscale
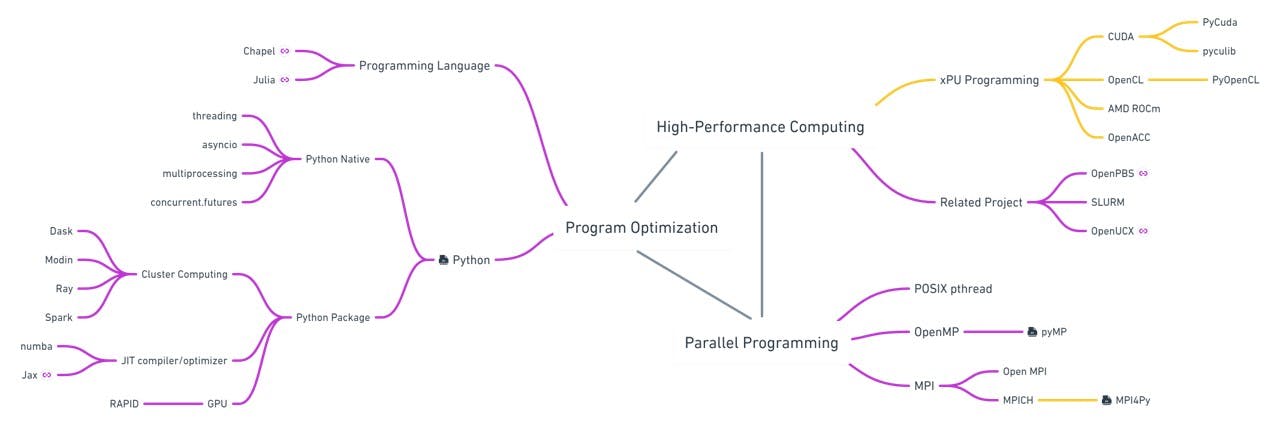
I have summarized related topics into a mind map. The world is constantly changing, if you have any other exciting projects feel free to comment below and I'll add them to the mind map as well.
Additional Reading:
Conclusion
It's not about which is the best. It's about which one is best for your situation. If you want to stick with pandas, try changing your code to take chunking. If you want to speed up your code, try exploring Dask as your processing engine. Try to understand the technology behind these packages and you'll know which one is right for your situation.
Additional Reading: